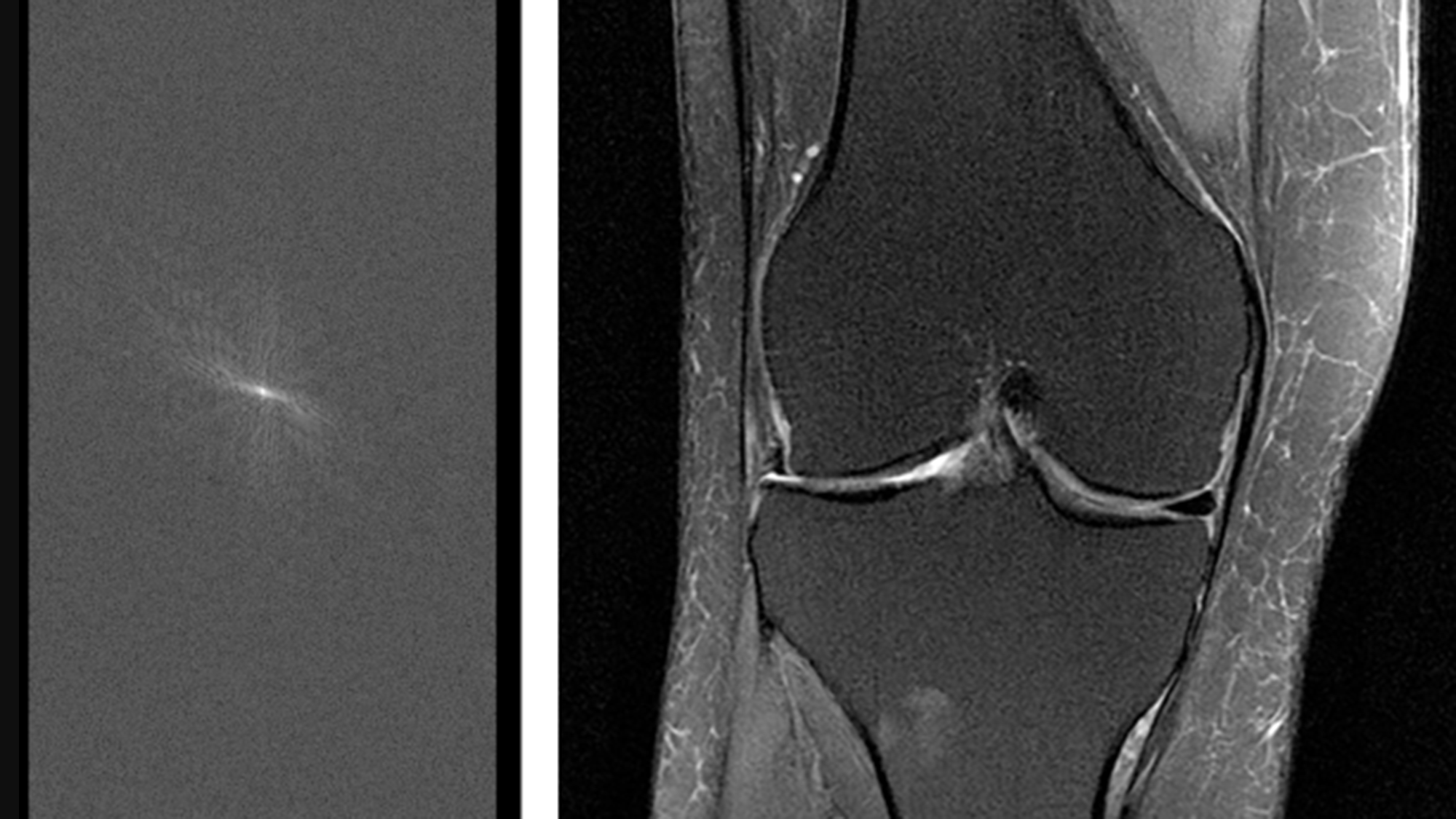
Facebook AI Research (FAIR) and NYU School of Medicine’s Center for Advanced Imaging Innovation and Research (CAI²R) are sharing new open source tools and data as part of fastMRI, a joint research project to spur development of AI systems to speed MRI scans by up to 10x. Today’s releases include new AI models and baselines for this task (as described in our paper here). It also includes the first large-scale MRI data set of its kind, which can serve as a benchmark for future research.
By sharing a standardized set of AI tools and MRI data, as well as hosting a leaderboard where research teams can compare their results, we aim to help improve diagnostic imaging technology, and eventually increase patients’ access to a powerful and sometimes life-saving technology. With new AI techniques, we hope to generate scans that require much less measurement data to produce the image detail necessary for accurate detection of abnormalities. Sharing this suite of resources reflects the fastMRI mission, which is to engage the larger community of AI and medical imaging researchers rather than to develop proprietary methods for accelerating MR imaging.
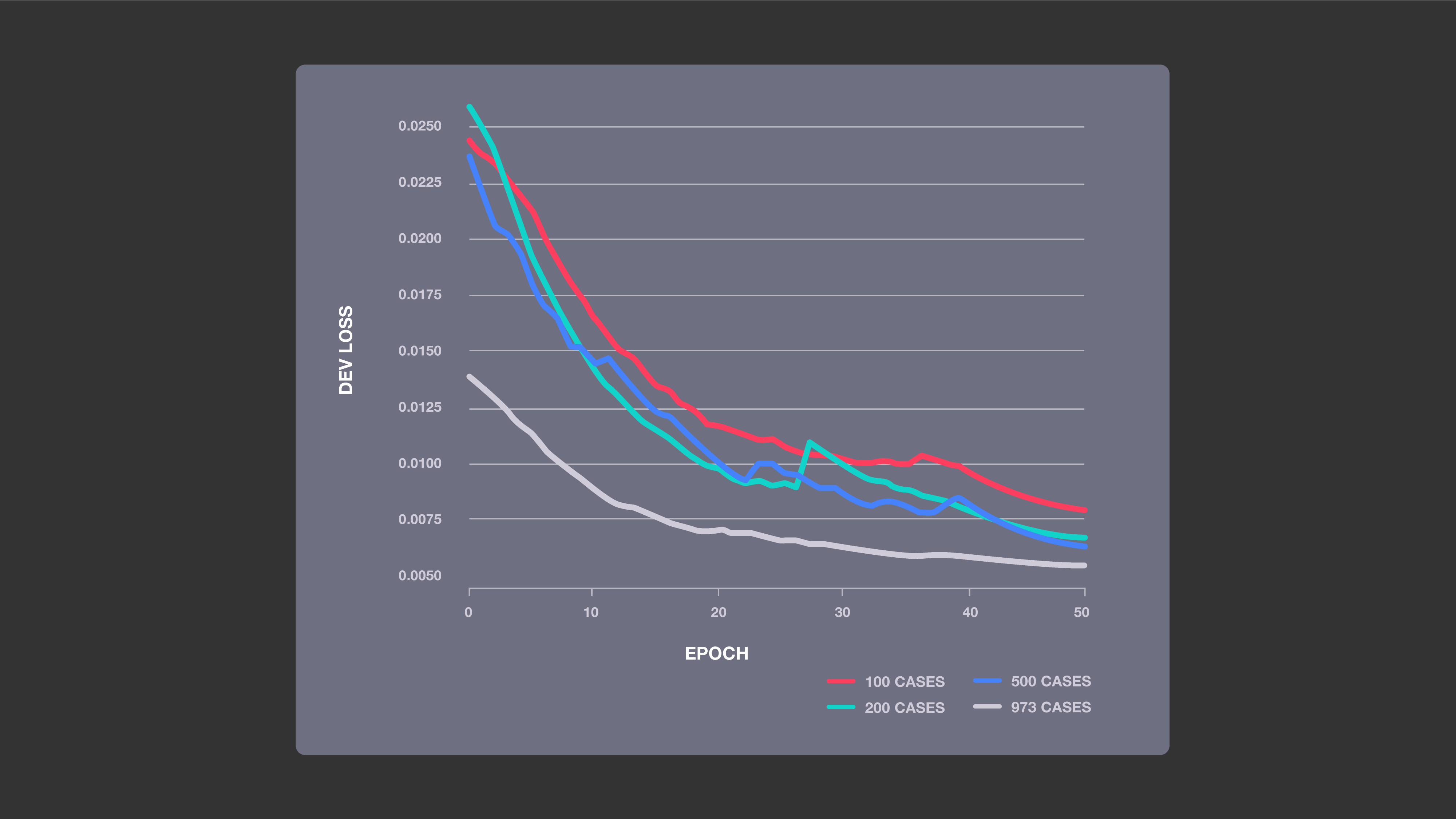
Improved accuracy with increased training data: This chart shows the impact of four sizes of training sets on the performance of our baseline ML models, which used a U-net neural network architecture. The more MRI cases the system was trained on, the lower its loss was, indicating that its image predictions were more likely to be accurate.
This work also helps us tackle fundamental research problems related to using AI to generate images from limited data. Unlike research in which neural networks create images that resemble the paintings, fashion designs, or other ground truth examples they were trained on, the AI-created images constructed from accelerated scans (which provide less data) must also be as accurate and detail-rich as standard MR images. Success in this difficult task could advance the state of AI while also making meaningful improvements in patient care.
The first large-scale database for reconstructing MRI scans
In the more than four decades since medical MR imaging was introduced, researchers have tried consistently to shorten the technology’s long scan times, which can sometimes require patients to remain stationary for more than an hour. But in 2016, research from the NYU School of Medicine showed that machine learning (ML) could significantly reduce scan durations by generating complete MR images from partial data. During a single exam, MRI devices collect a series of individual 2D spatial measurements — known as k-space data in the medical imaging community — and convert them into various images. By training neural networks on a large amounts of k-space data, this image reconstruction technique allows for less detailed initial scans, with the AI system generating complete images from a limited amount of data. This includes producing image details that might indicate a tumor, a ruptured blood vessel, or other key diagnostic feature.
The ultimate goal of the fastMRI project is to use AI-driven image reconstruction to achieve up to a 10x reduction in scan times. To begin, we’re providing baseline models for ML-based image reconstruction from k-space data subsampled at 4x and 8x scan accelerations. And we’ve already seen promising preliminary results for accelerating MR imaging by up to four times.
“We are excited that our preliminary work with acceleration factors up to four has shown interchangeability with conventional images. We hope that the release of this landmark data set, the largest-ever collection of fully sampled MRI raw data, will provide researchers with the tools necessary to achieve even greater acceleration factors,” says Michael P. Recht, M.D., Chair and the Louis Marx Professor of Radiology at NYU Langone Health.
One of the challenges facing the new research field of MR reconstruction is consistency, with teams using a variety of different data sets to train their AI systems. By releasing the largest open source database of MR data designed to tackle the problem of MR image reconstruction, NYU School of Medicine, a part of NYU Langone Health, hopes to provide an industry-wide and benchmark-ready data set. Just as the introduction of the widely used ImageNet data set advanced the state of computer vision research, the fastMRI data set could help organize and accelerate work related to MR reconstruction. This initial release includes approximately 1.5 million MR images drawn from 10,000 scans, as well as raw measurement data from nearly 1,600 scans. Like all the data used or released by the fastMRI project, this data set was gathered as part of a study approved by NYU Langone’s Internal Review Board. NYU fully anonymized the data set, including metadata and image content manually, inspecting each and every Digital Imaging and Communications in Medicine (DICOM) image for unexpected protected health information.
The raw measurement data in this data set sets it apart from previous MR databases and could prove particularly valuable for researchers. It consists of the k-space data that’s collected during scanning and typically discarded after it’s used to generate images. Instead, by retaining a large amount of raw k-space measurements and sharing them in an open source data set, NYU School of Medicine is providing researchers with unprecedented access to data for the purposes of training models, validating their performance, and generally simulating how image reconstruction techniques would be used in real-world conditions. The fastMRI data set also includes undersampled versions of those measurements, with k-space lines retrospectively masked, to simulate partial-data scans.
The k-space data in this data set is drawn from MR devices with multiple magnetic coils, and it also includes data that simulates measurements from single-coil machines. Though research related to accelerating multi-coil scans is more relevant for clinical practice — they’re more precise and more common than single-coil scans — the inclusion of single-coil k-space data measurements offers AI researchers an entry point for applying ML to this imaging task.
For additional information about the fastMRI data set and to request access, visit fastmri.med.nyu.edu.
Tools and a results leaderboard to help standardize MRI acceleration research
With NYU School of Medicine now providing the MR images and raw measurements, FAIR is sharing open source tools to make use of that data, while also ensuring that related work is reproducible and can be evaluated with consistency. To cover the widest array of research questions and results, we focused on two tasks — single-coil reconstruction and multi-coil reconstruction. For each task, we developed a baseline for classical, non-AI-based reconstruction methods and a separate baseline that incorporates deep learning models.
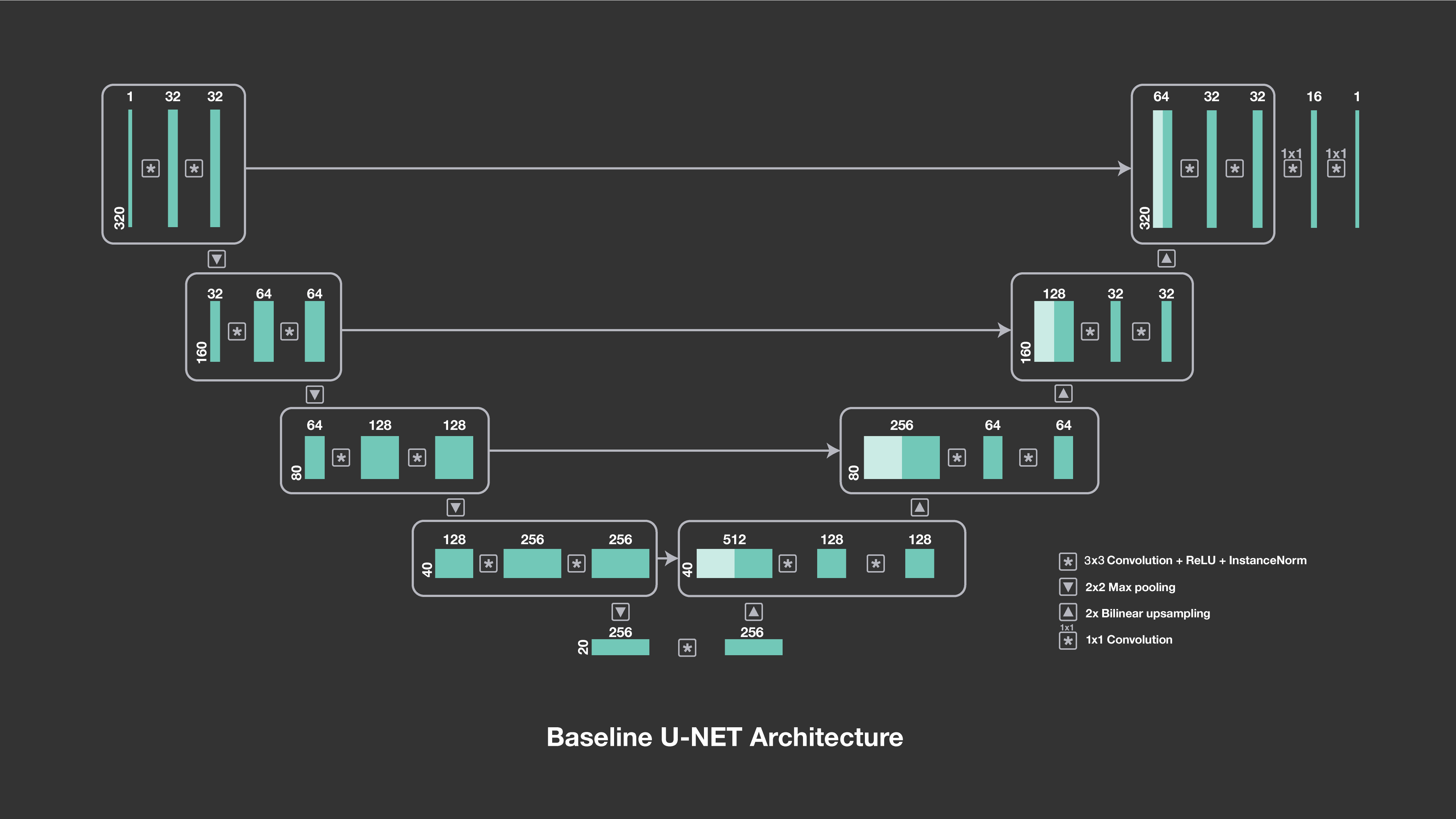
The u-net architecture consists of a downsampling path followed by an upsampling path. The numbers above the blocks indicate the number of channels, and the symbols show their interactions, including downsampling, upsampling and convolutions.
In both the single-coil and multi-coil deep learning baselines, our models are based on u-nets, a convolutional network architecture that was developed specifically for image segmentation in biomedical applications and that has a proven track record with image-to-image prediction. Our initial work has indicated that the u-net architecture is particularly responsive to training on a large amount of data, such as the MR reconstruction data set released by NYU School of Medicine.
To ensure the consistent measurement of progress within this field, FAIR has created a leaderboard for MR reconstruction results. We’ve added our baseline models to start, and other researchers will add improved results as they begin generating and submitting results to conferences and journals using the fastMRI data set. More than a competition, this leaderboard will allow the larger community of researchers to evaluate their results against consistent metrics and to see how different approaches compare. For more information about the leaderboard, and to submit results, visit fastMRI.org.
The next phase for fastMRI
Though its open source database is already larger than previous data sets released for MR reconstruction, NYU School of Medicine plans to release additional images and measurements in the near future, related to brain and liver scans (the current data set consists of knee scans). In addition, in the future FAIR and NYU School of Medicine plan to launch a reconstruction challenge, with participants producing results within a limited time frame (as opposed to the fastMRI leaderboard, where results will be posted on an ongoing basis). Winners of the challenge will be invited to present and share their work at a conference workshop. We’ll announce more details, including challenge timing and conference involvement in the coming months.
Our priority for the next phase of this collaboration is to use the experimental foundations we’ve established — the data and baselines — to further explore AI-based image reconstruction techniques. Additionally, any progress that we make at FAIR and NYU School of Medicine will be part of a larger collaboration that spans multiple research communities. We’re excited to see how others use our tools for this task and eager to compare our results on the fastMRI leaderboard, as well as in the research papers and workshops to come.